Parsing the Unstructured: Building a Gemini-Powered Prescription Parser in Go
Multi-modal LLMs like Gemini offer a powerful alternative to OCR for extracting structured data from unstructured docs. This post walks through a Go-based parser for prescriptions, using Gemini and RAG to produce high-accuracy JSON from handwritten and faxed forms.

Multi-modal LLMs like Google's Gemini are a powerful new alternative to traditional OCR for prescription parsing and other structured data extraction tasks. This makes Gemini ideal for transforming unstructured healthcare documents into clean JSON using Go. Healthcare documents like prescriptions are handwritten, scanned, and transmitted via fax, making them a challenge for traditional OCR.
To be fully transparent, I was fortunate enough to work with the Gemini team on a proof of concept of this use case while at a previous employer. While the PoC was promising, for various non-technical reasons, we never productized it. The code here and in the linked GitHub repository was produced specifically for this post. No code from that earlier PoC (which was C#) was used for this post or the linked GitHub project.
The full code of the prescription parser can be downloaded from this GitHub repository.
Identifying Real AI Use Cases (Not Just Chatbots)
In the past few years, it's seemed like AI is taking over the world. Most likely, you've been getting pressure to add AI to your software products to demonstrate that your company is embracing the future. Approaching AI this way, as a solution in search of a problem, has led many teams to add chatbots to their products with highly questionable value.
However, one use case that seems to have been largely overlooked is using LLMs in places where legacy OCR techniques either fall short or require significant effort to achieve high-quality results. Traditional OCR techniques usually require manual labeling and a consistent input format to get consistently high-quality output.
Why LLMs Beat Traditional OCR for Prescription Parsing
The design of traditional OCR is particularly challenging when dealing with input like prescription images. You may have significantly different manufacturer or drug-specific referral forms that use different fields for the same data. Add in the fact that these forms may be hand-filled and then transmitted over fax, and the difficulties just compound.
Traditional OCR can overcome all of these challenges, but the effort required to tune and maintain these models can be significant. High-accuracy structured data extraction with traditional OCR can require a large volume of sample data that's been manually labeled, specifying field locations on the input images. You'll need a large enough sample for training and an evaluation dataset to assess model performance and conduct fine-tuning.
Effectively handling data variations may require building a domain-specific taxonomy to normalize the output correctly. If the input image structure changes significantly (a new manufacturer/drug, for example), the model performance will dramatically degrade and require retraining your model.
Using LLMs for Prescription Parsing and Structured Data Extraction
LLM-based data extraction can overcome all these challenges without creating fine-tuned models. The design of LLMs naturally allows them to apply strong semantic reasoning to normalize input data variations without needing specific instructions. This semantic reasoning and the large volume of data foundation models are trained on also means they may already have a strong domain-specific knowledge base without fine-tuning. Finally, LLMs are a natural fit for converting raw text into structured formats like JSON.
Coupling LLM-based document extraction with RAG (retrieval augmented generation) also allows for continuous reinforcement feedback loops without running full fine-tuning training cycles. By generating and storing embeddings from validated examples, we can perform semantic search to find the most relevant samples to include in context when parsing new documents.
Google's Gemini's Industry-Leading Multimodal Capabilities
Companies like OpenAI and Anthropic tend to dominate the news cycles regarding LLMs. However, when it comes to multi-modal LLMs, I feel like Google has been quietly dominating with Gemini for years now. I initially built this prescription parser in Go using OpenAI to test its multimodal capabilities, but Gemini delivered significantly better results for healthcare document parsing. So, I updated the code to support both OpenAI and Gemini as parser backends.
Because Gemini produced consistently better results, and I recently released another blog post about using the OpenAI responses API in Go, this blog post will focus only on Gemini. Feel free to review the full code on GitHub to see the OpenAI parser implementation.
Architecture Overview: Multi-Pass Parsing with Gemini and RAG
Our prescription parser flow will be designed as a multiple-pass parser to get the highest accuracy and output normalization without fine-tuning. The parser service will support endpoints for parsing prescriptions and ingesting validated samples for our feedback loop.
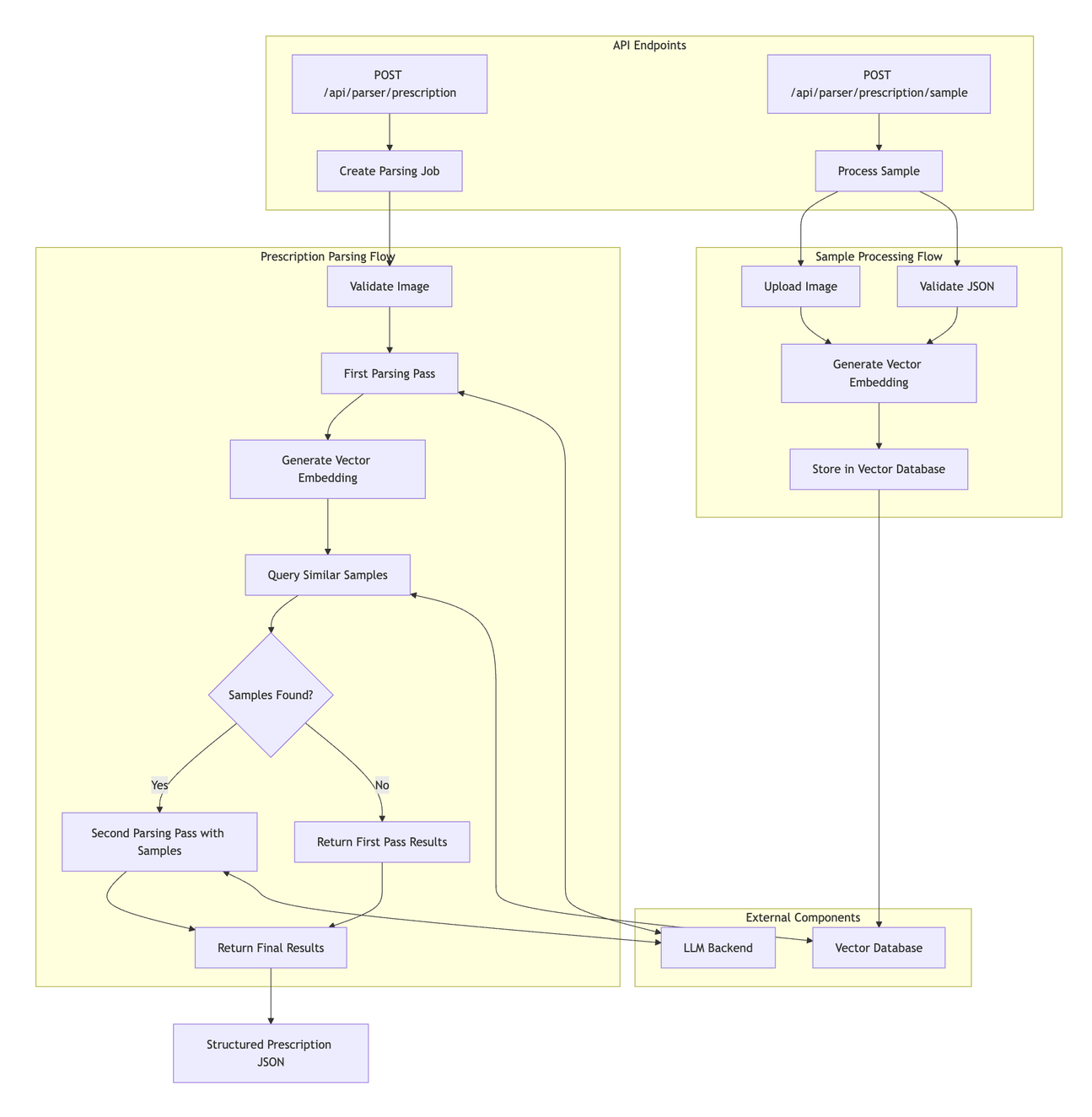
A POST request containing a prescription image and its corresponding JSON representation will trigger the feedback flow. The parser service will save the prescription image to persistent storage, generate a vector embedding of the JSON, and store the JSON and the embedding in the database.
A POST request containing a prescription image will trigger the parsing flow. The parser service will perform a first pass to get an initial JSON representation of the prescription. The service will generate an embedding from the initial prescription JSON to perform a semantic search against the vector database to find the most relevant sample prescriptions. The parser service will then perform a second parsing pass to review the first pass JSON output with the sample prescriptions in context.
Implementing the Prescription Parser
Let’s walk through how this Go-based prescription parsing system works, from ingesting scanned images to producing clean JSON using Gemini’s multimodal LLM API.
The API Handler
Because our multi-pass parsing process can take a long time, we'll design it to be asynchronous so that client requests don't have to wait for the full parsing to complete. The parsing service will support two endpoints for the prescription parsing flow. One endpoint will accept a prescription image payload and immediately return a job ID after launching a background goroutine for parsing. The second endpoint will allow clients to request the status of a parsing job by ID. When the job status is complete the response will include a result field with the prescription JSON.
// RegisterRoutes registers all parser-related routes
func (h *Handler) RegisterRoutes(router *mux.Router) {
parserRouter := router.PathPrefix("/parser").Subrouter()
parserRouter.HandleFunc("/prescription", h.ParsePrescription).Methods("POST")
parserRouter.HandleFunc("/prescription/sample", h.SaveSamplePrescription).Methods("POST")
parserRouter.HandleFunc("/prescription/{id}", h.GetJobStatus).Methods("GET")
}Generating Structured Data from Documents using Gemini
Our parsing flow is broken up into three steps: first parsing pass, sample lookup, and assuming we have samples, a second parsing pass.
The first parsing pass is straightforward; we provide system instructions on how to parse a prescription, the JSON schema for our structured output, and the image file along with a brief prompt instructing the model to parse the image into our JSON format.
// firstParsingPass performs the initial parsing of the prescription.
// It sends the prescription image to OpenAI API with system and user prompts
// to extract structured data from the image.
func (p *OpenAIParser) firstParsingPass(ctx context.Context, fileID string) (models.Prescription, error) {
messages := []responses.ResponseInputItemUnionParam{
responses.ResponseInputItemParamOfMessage(
systemPrompt,
"system"),
}
imageMessage := responses.ResponseInputItemParamOfMessage(
responses.ResponseInputMessageContentListParam{
responses.ResponseInputContentUnionParam{
OfInputFile: &responses.ResponseInputFileParam{
FileID: openai.String(fileID),
Type: "input_file",
},
},
responses.ResponseInputContentUnionParam{
OfInputText: &responses.ResponseInputTextParam{
Text: parsePrompt,
Type: "input_text",
},
},
},
"user",
)
messages = append(messages, imageMessage)
params := responses.ResponseNewParams{
Text: responses.ResponseTextConfigParam{
Format: responses.ResponseFormatTextConfigUnionParam{
OfJSONSchema: &responses.ResponseFormatTextJSONSchemaConfigParam{
Name: "Prescription",
Schema: PrescriptionResponseSchema,
Strict: openai.Bool(true),
Description: openai.String("Prescription Image Parser Prescription JSON"),
Type: "json_schema",
},
},
},
Model: "gpt-4.1-2025-04-14",
Input: responses.ResponseNewParamsInputUnion{
OfInputItemList: messages,
},
MaxOutputTokens: openai.Int(10240),
}
resp, err := p.client.Responses.New(ctx, params)
if err != nil {
return models.Prescription{}, fmt.Errorf("failed to process image: %w", err)
}
var rx models.Prescription
err = json.Unmarshal([]byte(resp.OutputText()), &rx)
if err != nil {
return models.Prescription{}, fmt.Errorf("failed to unmarshal response: %w", err)
}
return rx, nil
}Once we have a JSON representation of the prescription, we'll generate embeddings and call our datastore method to get related sample prescriptions using the embeddings.
// Get embedding for the parsed prescription
embedding, err := p.GetEmbedding(ctx, rx)
if err != nil {
p.logger.Error("failed to get embedding", zap.String("job_id", jobID), zap.String("file_name", fileName), zap.Error(err))
jobs.GlobalTracker.UpdateJob(jobID, jobs.JobStatusComplete, nil, rx)
return
}
// Get similar samples
samples, err := p.ds.GetSamples(ctx, embedding)
if err != nil {
p.logger.Error("failed to get samples", zap.String("job_id", jobID), zap.String("file_name", fileName), zap.Error(err))
jobs.GlobalTracker.UpdateJob(jobID, jobs.JobStatusComplete, nil, rx)
return
}
p.logger.Info("sample images loaded", zap.String("job_id", jobID), zap.String("file_name", fileName), zap.Int("sample_count", len(samples)))
// Second parsing pass with examples
if len(samples) > 0 {
secondPassRx, err := p.secondParsingPass(ctx, storedFile.ID, samples, rx)
if err != nil {
p.logger.Error("failed in second parsing pass", zap.String("job_id", jobID), zap.Error(err))
jobs.GlobalTracker.UpdateJob(jobID, jobs.JobStatusComplete, nil, rx)
return
}
rx = secondPassRx
}
p.logger.Info("successfully processed image", zap.String("job_id", jobID), zap.String("file_name", fileName))
jobs.GlobalTracker.UpdateJob(jobID, jobs.JobStatusComplete, nil, rx)If the datastore returned sample prescriptions, we'll perform a second parsing pass to review the initial JSON with the sample prescriptions in context as message history.
// secondParsingPass performs a review with example context.
// It uses similar prescription samples to refine the initial parsing results,
// potentially improving accuracy by learning from precedents.
func (p *OpenAIParser) secondParsingPass(ctx context.Context, fileID string, samples []models.SamplePrescription, firstPassRx models.Prescription) (models.Prescription, error) {
messages := []responses.ResponseInputItemUnionParam{
responses.ResponseInputItemParamOfMessage(
systemPrompt,
"system"),
}
for _, sample := range samples {
sampleMessage := responses.ResponseInputItemParamOfMessage(
responses.ResponseInputMessageContentListParam{
responses.ResponseInputContentUnionParam{
OfInputFile: &responses.ResponseInputFileParam{
FileID: openai.String(sample.FileID),
Type: "input_file",
},
},
responses.ResponseInputContentUnionParam{
OfInputText: &responses.ResponseInputTextParam{
Text: parsePrompt,
Type: "input_text",
},
},
},
"user",
)
sampleResponse := responses.ResponseInputItemParamOfMessage(
responses.ResponseInputMessageContentListParam{
responses.ResponseInputContentUnionParam{
OfInputText: &responses.ResponseInputTextParam{
Text: sample.Content,
Type: "output_text",
},
},
},
"assistant",
)
messages = append(messages, sampleMessage)
messages = append(messages, sampleResponse)
}
imageMessage := responses.ResponseInputItemParamOfMessage(
responses.ResponseInputMessageContentListParam{
responses.ResponseInputContentUnionParam{
OfInputFile: &responses.ResponseInputFileParam{
FileID: openai.String(fileID),
Type: "input_file",
},
},
responses.ResponseInputContentUnionParam{
OfInputText: &responses.ResponseInputTextParam{
Text: parsePrompt,
Type: "input_text",
},
},
},
"user",
)
messages = append(messages, imageMessage)
firstPassResponseText, err := json.Marshal(firstPassRx)
if err != nil {
return firstPassRx, fmt.Errorf("failed to marshal first pass response: %w", err)
}
firstPassResponse := responses.ResponseInputItemParamOfMessage(
responses.ResponseInputMessageContentListParam{
responses.ResponseInputContentUnionParam{
OfInputText: &responses.ResponseInputTextParam{
Text: string(firstPassResponseText),
Type: "output_text",
},
},
},
"assistant",
)
messages = append(messages, firstPassResponse)
messages = append(
messages,
responses.ResponseInputItemParamOfMessage(
reviewPrompt,
"user",
),
)
params := responses.ResponseNewParams{
Text: responses.ResponseTextConfigParam{
Format: responses.ResponseFormatTextConfigUnionParam{
OfJSONSchema: &responses.ResponseFormatTextJSONSchemaConfigParam{
Name: "Prescription",
Schema: PrescriptionResponseSchema,
Strict: openai.Bool(true),
Description: openai.String("Prescription Image Parser Prescription JSON"),
Type: "json_schema",
},
},
},
Model: "gpt-4.1-2025-04-14",
Input: responses.ResponseNewParamsInputUnion{
OfInputItemList: messages,
},
MaxOutputTokens: openai.Int(10240),
}
resp, err := p.client.Responses.New(ctx, params)
if err != nil {
return firstPassRx, fmt.Errorf("failed to run second pass: %w", err)
}
var secondPassRx models.Prescription
err = json.Unmarshal([]byte(resp.OutputText()), &secondPassRx)
if err != nil {
return firstPassRx, fmt.Errorf("failed to unmarshal second pass response: %w", err)
}
return secondPassRx, nil
}Retrieval Augmented Generation (RAG) Using Go and Postgres
In addition to our actual prescription parser service and Google Gemini, we'll also use a vector database to store sample prescriptions and perform vector search to find the most relevant samples to augment our parser prompt. In this implementation, we'll use a Postgres database with the pgvector extension for performing semantic search.
The vector search implementation for our parser service is heavily based on this blog post. I've been using Ent as my first-choice ORM for Go for years now, so it was convenient that the project team recently published a blog about building a RAG with Go and pgvector.
We'll define two tables for our database, one for storing the sample prescriptions and one for storing the generated vector embeddings. The embeddings table will have a foreign key relationship with our prescriptions table allowing us to retrieve the full prescription JSON to feed into our LLM context based on the embeddings rows that match the semantic search.
// Prescription holds the schema definition for the Prescription entity.
type Prescription struct {
ent.Schema
}
// Fields of the Prescription.
func (Prescription) Fields() []ent.Field {
return []ent.Field{
field.UUID("id", uuid.UUID{}).
Default(uuid.New).
Immutable(),
field.String("file_id"),
field.String("mime_type"),
field.JSON("content", models.Prescription{}),
}
}
// Embedding holds the schema definition for the Embedding entity.
type Embedding struct {
ent.Schema
}
// Fields of the Embedding.
func (Embedding) Fields() []ent.Field {
return []ent.Field{
field.UUID("id", uuid.UUID{}).
Default(uuid.New).
Immutable(),
field.Other("embedding", pgvector.Vector{}).
SchemaType(map[string]string{
dialect.Postgres: "vector(1536)",
}),
}
}
// Edges of the Embedding.
func (Embedding) Edges() []ent.Edge {
return []ent.Edge{
edge.To("prescription", Prescription.Type).Unique().Required(),
}
}
func (Embedding) Indexes() []ent.Index {
return []ent.Index{
index.Fields("embedding").
Annotations(
entsql.IndexType("hnsw"),
entsql.OpClass("vector_l2_ops"),
),
}
}We'll add two methods to our datastore, one to save new prescription samples and one to perform a semantic search to find the samples that are the closest match for a given input prescription.
Because embeddings are specific to the LLM that we're using as our parser backend, we'll generate the embeddings independent of the actual datastore code.
Generating Embeddings with Gemini
We'll add a GetEmbedding method to our Parser implementation to use the Gemini API to generate embeddings from the prescription JSON.
// GetEmbedding generates embeddings for a prescription using OpenAI.
// It converts the prescription to JSON and sends it to the OpenAI API to generate
// a vector representation for similarity search.
func (p *OpenAIParser) GetEmbedding(ctx context.Context, prescription models.Prescription) ([]float32, error) {
jsonBytes, err := json.Marshal(prescription)
if err != nil {
return nil, fmt.Errorf("failed to marshal prescription: %w", err)
}
resp, err := p.client.Embeddings.New(ctx, openai.EmbeddingNewParams{
Input: openai.EmbeddingNewParamsInputUnion{
OfString: openai.String(string(jsonBytes)),
},
Model: openai.EmbeddingModelTextEmbedding3Small,
Dimensions: openai.Int(1536),
EncodingFormat: "float",
})
if err != nil || len(resp.Data) == 0 {
return nil, fmt.Errorf("failed to generate prescription embedding: %w", err)
}
var emb openAIEmbedding
err = json.Unmarshal([]byte(resp.Data[0].RawJSON()), &emb)
if err != nil {
return nil, fmt.Errorf("failed to unmarshal prescription embedding: %w", err)
}
return emb.Embedding, nil
}Indexing Documents for Retrieval
The SaveSamplePrescription method will accept the input prescription JSON, file details, and pre-generated embeddings and save them to the database.
// SaveSamplePrescription stores a prescription and its vector embedding in the database.
// It creates both the prescription record and its associated embedding in a single transaction.
//
// Parameters:
// - ctx: Context for the database operation
// - mimeType: MIME type of the prescription image
// - imageID: ID of the image file associated with this prescription
// - prescription: Prescription data to store
// - embedding: Vector embedding representing the prescription content for similarity search
//
// Returns:
// - An error if the database operation fails, nil on success
func (d *PgEntDatastore) SaveSamplePrescription(ctx context.Context, mimeType, imageID string, prescription models.Prescription, embedding []float32) error {
tx, err := d.dbClient.Tx(ctx)
if err != nil {
d.logger.Error("failed to create transaction", zap.Error(err))
return fmt.Errorf("failed to create transaction: %w", err)
}
defer tx.Rollback()
dbPrescription, err := tx.Prescription.Create().
SetFileID(imageID).
SetMimeType(mimeType).
SetContent(prescription).
Save(ctx)
if err != nil {
d.logger.Error("failed to create prescription", zap.Error(err))
return fmt.Errorf("failed to create prescription: %w", err)
}
_, err = tx.Embedding.Create().
SetPrescriptionID(dbPrescription.ID).
SetEmbedding(pgvector.NewVector(embedding)).
Save(ctx)
if err != nil {
d.logger.Error("failed to create embedding", zap.Error(err))
return fmt.Errorf("failed to create embedding: %w", err)
}
err = tx.Commit()
if err != nil {
d.logger.Error("failed to commit transaction", zap.Error(err))
return fmt.Errorf("failed to commit transaction: %w", err)
}
return nil
}
Performing Vector Search with Go using pgvector
The GetSamples method will accept the pre-generated embeddings for a new prescription, perform a semantic search against the vector store, and return a slice of the closest matching sample prescriptions.
// GetSamples retrieves prescription samples that are most similar to the provided embedding vector.
// It uses pgvector's similarity search to find the closest matches in the embedding space.
// The method returns up to 3 most similar samples, ordered by vector similarity.
//
// Parameters:
// - ctx: Context for the database operation
// - embedding: Vector embedding to use for similarity search
//
// Returns:
// - A slice of SamplePrescription that are most similar to the embedding
// - An error if the database operation fails
func (d *PgEntDatastore) GetSamples(ctx context.Context, embedding []float32) ([]models.SamplePrescription, error) {
var samples []models.SamplePrescription
tx, err := d.dbClient.BeginTx(ctx, nil)
if err != nil {
return nil, fmt.Errorf("failed to start transaction: %w", err)
}
defer tx.Rollback()
embVec := pgvector.NewVector(embedding)
embs, err := tx.Embedding.Query().
Order(func(s *sql.Selector) {
s.OrderExpr(sql.ExprP("embedding <-> $1", embVec))
}).
WithPrescription().
Limit(3).
All(ctx)
if err != nil {
return nil, fmt.Errorf("failed to get samples: %w", err)
}
for _, emb := range embs {
if emb.Edges.Prescription != nil {
content, err := json.Marshal(emb.Edges.Prescription.Content)
if err != nil {
d.logger.Error("failed to marshal prescription content", zap.Error(err))
continue
}
samples = append(samples, models.SamplePrescription{
ID: emb.Edges.Prescription.ID,
FileID: emb.Edges.Prescription.FileID,
MIMEType: emb.Edges.Prescription.MimeType,
Content: string(content),
})
}
}
if err := tx.Commit(); err != nil {
return nil, fmt.Errorf("failed to commit transaction: %w", err)
}
return samples, nil
}Gemini Output Example: Structured JSON from a Handwritten Prescription
The multi-modal capabilities of the Gemini models have advanced so significantly that out of the box, it can extract prescription data from an image with an average of 95% accuracy.
go run ./cmd/parser-eval -pdf ./samples/Humira4.pdf -json ./samples/Humira4.json
Filename: Humira4.pdf - Job ID: b1d1f903-5429-4d04-ba7f-6b05472d30f8
Score: 95.83% - (69.00 / 72)
Feedback:
The parser demonstrated high accuracy across most fields, achieving exact matches for a significant portion of the data. Minor semantic differences were observed in the 'clinical_info' and 'medications[0].administration_notes' fields, which were scored as semantically equivalent. However, two fields, 'patient.phone_numbers[0].label' and 'prescriber.npi', showed significant discrepancies, and most notably, the 'medications[0].indication' field was completely missed by the parser, resulting in a 0.0 score. Overall, the parser performed very well, but there are clear areas for improvement regarding specific data points and ensuring completeness.Here's an example of the parser output for a handwritten (fictitious) prescription image.
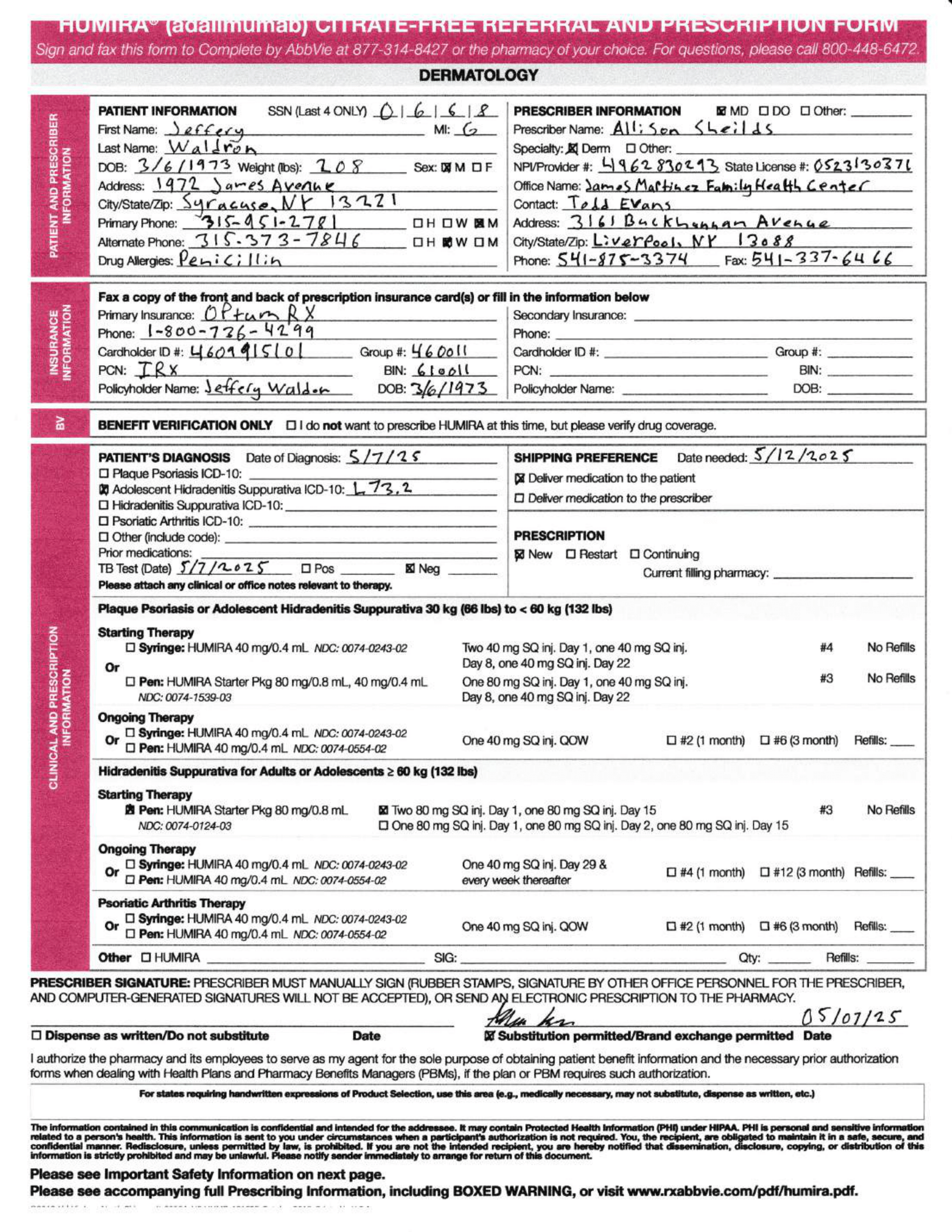
Here's the JSON output for the prescription:
{
"date_written": "2025-05-07",
"date_needed": "",
"patient": {
"first_name": "Jeffery",
"middle_name": "G",
"last_name": "Waldron",
"dob": "1973-03-06",
"sex": "Male",
"weight": {
"unit": "lbs",
"value": "208"
},
"height": {
"unit": "",
"value": ""
},
"address": {
"street": "1972 James Avenue",
"city": "Syracuse",
"state": "NY",
"zip": "13221"
},
"phone_numbers": [
{
"label": "Home",
"number": "3159512781",
"extension": ""
},
{
"label": "Alternate",
"number": "3153737846",
"extension": ""
}
],
"allergies": [
"Penicillin"
],
"emergency_contact": {
"name": "",
"relationship": "",
"phone": ""
},
"insurance": [
{
"type": "Primary",
"provider": "Optum RX",
"id_number": "4609915101",
"group_number": "460011",
"rx_bin": "610011",
"pcn": "IRX",
"policyholder_name": "Jeffery Waldron",
"policyholder_dob": "1973-03-06",
"phone_number": "18007364299"
}
]
},
"prescriber": {
"name": "Allison Sheilds",
"specialty": "Dermatology",
"npi": "4962830293",
"state_license": "0523130371",
"dea": "",
"office": {
"name": "James Martinez Family Health Center",
"address": {
"street": "3161 Buckhannan Avenue",
"city": "Liverpool",
"state": "NY",
"zip": "13088"
},
"phone": "5418753374",
"fax": "5413376466",
"contact_name": "Todd Evans",
"contact_email": ""
}
},
"diagnosis": {
"date_of_diagnosis": "2025-05-07",
"primary_diagnosis": {
"description": "Adolescent Hidradenitis Suppurativa",
"icd10_code": "L73.2"
},
"additional_diagnoses": null
},
"clinical_info": [
"TB Test Date: 2025-05-07, Result: Negative"
],
"medications": [
{
"drug_name": "HUMIRA (adalimumab) Starter Pkg",
"ndc": "0074-0124-03",
"form": "Pen",
"strength": "80 mg/0.8 mL",
"sig": "Two 80 mg SQ inj. Day 1, one 80 mg SQ inj. Day 15",
"quantity": "3",
"refills": "0",
"start_date": "",
"duration": "",
"administration_notes": "Inject two 80 mg subcutaneously on Day 1, then inject one 80 mg subcutaneously on Day 15.",
"indication": "Hidradenitis Suppurativa"
}
],
"therapy_status": "New",
"failed_therapies": null,
"delivery": {
"destination": "Patient's Home",
"notes": ""
},
"prescriber_signature": {
"date": "2025-05-07",
"daw_code": "0"
},
"attachments": {
"insurance_cards": false,
"lab_results": false,
"pathology_reports": false,
"clinical_notes": false,
"other_documents": false
}
}Why Prompt Testing Matters for Production Reliability
As you can see, the code required to build a prescription parser using Gemini is pretty straightforward. It's the prompt design itself that adds a lot of complexity. Adding or changing instructions related to one prescription attribute can have a surprising negative impact on parsing an entirely different prescription attribute.
It's critical when building LLM-based applications like this to create test harnesses that can take known inputs and their expected outputs and score the results of the extraction to allow you to catch regressions in output quality as you adjust prompts.
I haven't added a test harness like that to this project, but I plan on adding one. Having a harness like this is also invaluable for comparing performance when evaluating different models.
Wrapping Up: Practical Applications for AI
Hopefully, we're past the initial gold rush of cramming AI into existing applications, when everyone was blindly adding chatbots to their apps. That said, the pressure to integrate AI into your applications (and your development process) will only increase. One area where I think LLMs, especially powerful multimodal models like Gemini, have real, underutilized value is in use cases like prescription parsing and healthcare structured data extraction.
Have you used LLMs for structured data extraction? Do you have other use cases where you've successfully integrated AI into your products? I'd love to hear your thoughts and feedback in the comments.


